Plot processes¶
The WPS flyingpigeon provides several processes to perform plot subsetts of netCDF files.
They are several processes to perfom timeseries graphics as well as spatial visualisations as maps.
[1]:
# birdy client for communication with the server:
from birdy import WPSClient
from os import environ
# handling files and folders
from os import path, listdir
from urllib import request
# wait until WPS process is finished
import time
# to display external png graphics in notebook:
from IPython.display import Image
from IPython.core.display import HTML
[2]:
# This cell is for server admnistration test purpose
# Ignore this cell and modify the following cell according to your needs
fp_server = environ.get('FYINGPIGEON_WPS_URL')
print(fp_server) # link to the flyingpigoen server
http://localhost:8093/wps
[3]:
# URL to a flyingpigeon server
# fp_server = 'https://pavics.ouranos.ca/twitcher/ows/proxy/flyginpigeon/wps'
[4]:
fp_i = WPSClient(fp_server, progress=True)
fp = WPSClient(fp_server)
read in the required files:
this is an example of an local installation, where local files are processed
indices were calculated with the birdhouse WPS finch: https://finch.readthedocs.io/en/latest/processes.html
[5]:
# read in the existing indices based on bias_adjusted tas files:
tas_NER = '/home/nils/data/example_data/NER/'
tasInd_NER = [ tas_NER+f for f in listdir(tas_NER) if '.nc' in f ]
tasInd_NER.sort()
[ ]:
pip install pymetalink
[6]:
# frequencies
freq=['yr','mon']
# precipitation indices
# pr_indices = ['prcptot','rx1day','wetdays','cdd','cwd','sdii','rx5day']
tas_indice = 'tg_mean'
# titles = ['Somme annuelle des précipitations',
# 'Jours de plus fortes précipitations',
# 'Nombre de jours humide',
# 'Journées consécutives de sécheresse',
# 'Jours humides consécutifs',
# "Index d'intensité de précipitations",
# 'Somme max. sur 5 jours consécutifs',
# 'Températures moyennes annuelles']
dates = ['1976-01-01', '2005-12-31', '2036-01-01', '2065-12-31', '2071-01-01', '2099-12-30']
[7]:
# find ensemble files for one indice based on pr files:
tasInd_NER = [ tas_NER+f for f in listdir(tas_NER) if '.nc' in f ]
resource = [f for f in tasInd_NER if tas_indice in f ] # and '_yr_' in f
Spaghetti Plot¶
A simple way of visualisation of an ensemble of datasets. The plot visualises historical and rcp runs in different colors
[8]:
out_plot = fp.plot_spaghetti(resource=resource, title='Test plot',
delta = -273.15, # to convert K to C
figsize='9,5',# ymin=0, ymax=14 #
)
len(out_plot.get())
[8]:
1
[9]:
# display the output graphic url:
out_plot.get(asobj=True).plotout_spaghetti
# or with:
# Image(out.get()[0], width=400)
[9]:
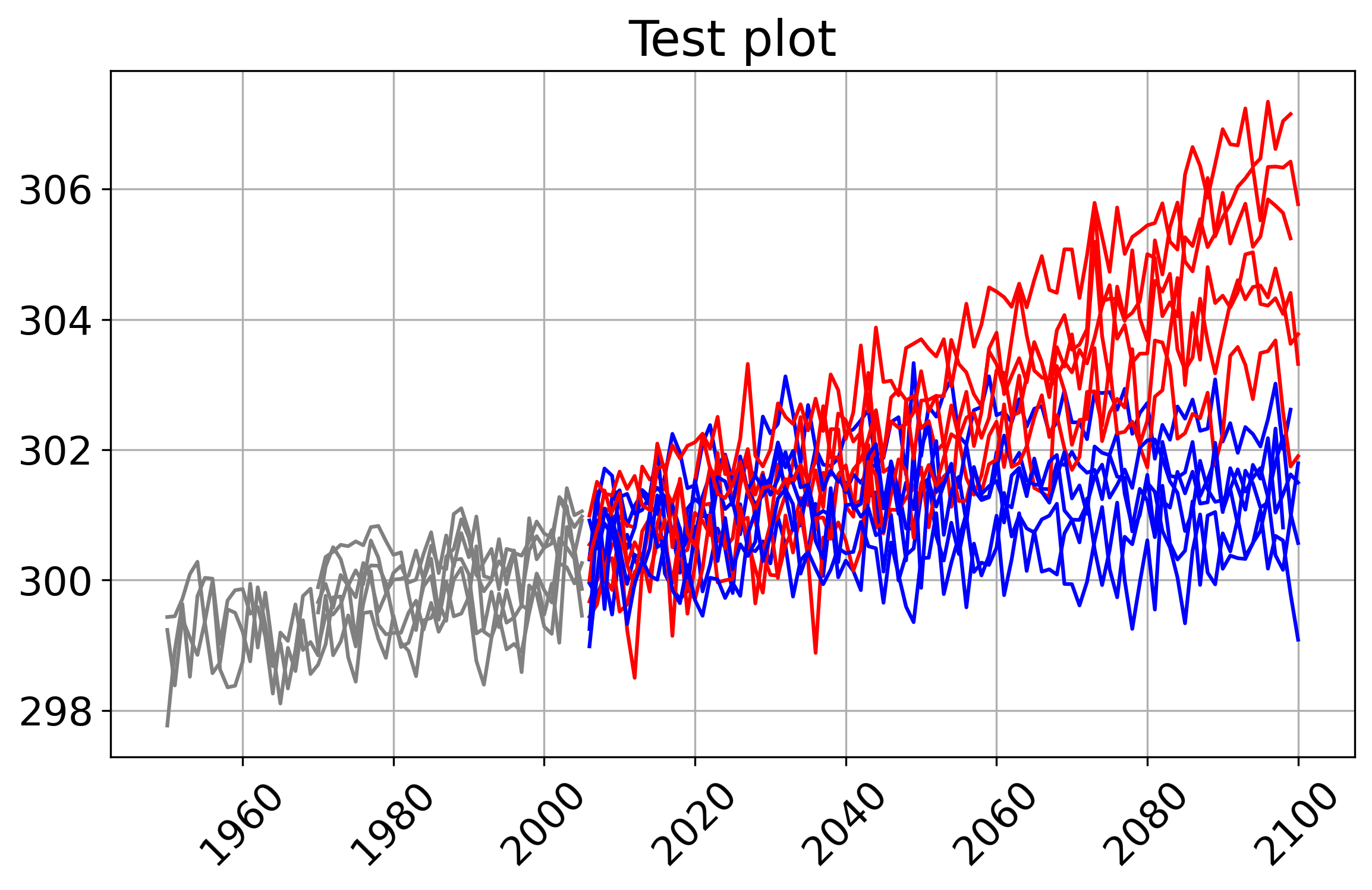
RCP uncertainty timeseries¶
An ensemble of indice files will be visualised as median and the appropriate uncertainties seperatled by historical and rcp runs.
[10]:
resp = fp.plot_uncertaintyrcp(resource=resource, title='Yearly mean temperature', delta = -273.15,
figsize='9,5', # ymin=20, ymax=100 #
)
[11]:
# onother way to display the output graphic.
# download the file
out_file = '/tmp/ts_uncertainty_tg-yr.png'
request.urlretrieve(resp.get()[0], out_file)
# display the graphic:
Image(out_file, width=400)
[11]:

Plot a map¶
Spatial visualisation of data as a mean over the time.
[12]:
# select only the historical runs of the indices ensemble
hist = [f for f in resource if 'historical' in f]
# process the data of the years 1971-2000
out = fp.plot_map_timemean(resource=hist, title='Historical reference (1971-2000)',
delta = -273.15,
datestart='1971-01-01', dateend='2000-12-31', # subset a time range
vmin=20, vmax=30,
# cmap=''
) #
# display the output graphic
Image(out.get()[0], width=600)
[12]:
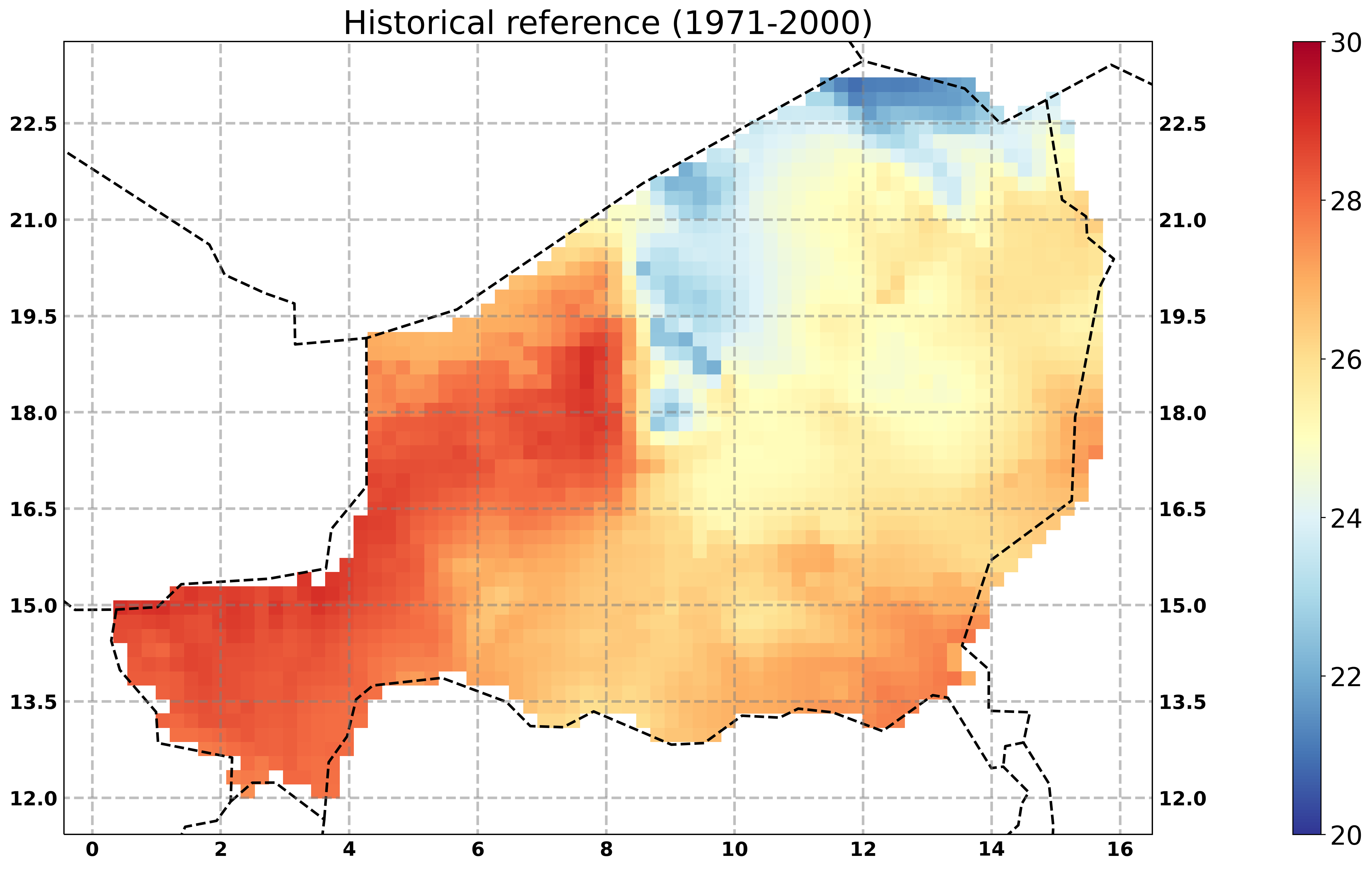
plot a climate change signal¶
calculation of the differnence in a climate signal between a future projection to a reference periode
[13]:
dates = ['1971-01-01', '2000-12-31', '2036-01-01', '2065-12-31', '2071-01-01', '2100-12-31']
ref = [f for f in tasInd_NER if tas_indice in f and 'historical' in f] # and fr in f
proj = [f for f in tasInd_NER if tas_indice in f and 'rcp85' in f]
ref.sort()
proj.sort()
[14]:
resp = fp_i.climatechange_signal(resource_ref=ref,
resource_proj=proj,
# variable='tg-mean',
title='Scenario {} {} ({})'.format('RCP 8.5', tas_indice, '2071-2100'),
datestart_ref=dates[0],
dateend_ref=dates[1],
datestart_proj=dates[2],
dateend_proj=dates[3],
# vmin=0 , vmax=7,
# cmap='BrBG'
)
[15]:
# using fp_i you need to wait until the processing is complete!
timeout = time.time() + 60*1 # 1 minute from now
while resp.getStatus() != 'ProcessSucceeded':
time.sleep(1)
if time.time() > timeout: # to avoid endless waiting if the process failed
break
Image(resp.get()[2], width=600)
[15]:
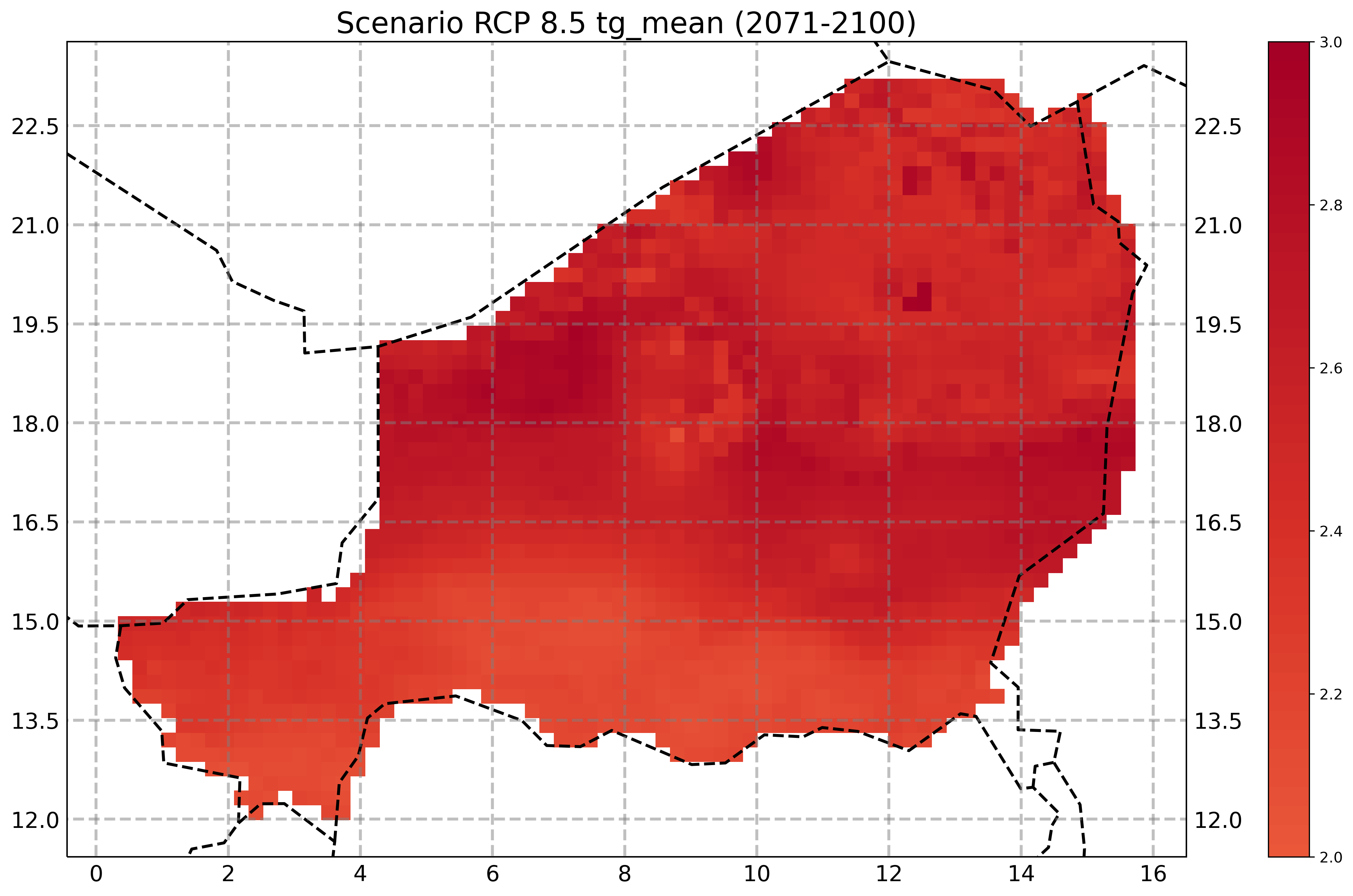
[ ]: