Subset processes¶
The WPS flyingpigeon provides several processes to perform spatial subsets of netCDF files:
subset_bbox: Crop netCDF files to a given latitude longitude bounding box.
subset_countries: Crop netCDF files to only the intersection of predefined countries.
subset_continents: Crop netCDF files to only the intersection of predefined continents.
subset_wfs_polygon: Crop netCDF files to only the intersection of given polygons available on a given WFS server.
Note that subsetting operations do not average or reduce results over the selected area, they only return the grid sells intersecting with the area. A mask is applied to cells that are outside the selected area, but inside the rectangular grid required for netCDF files.
[1]:
# Import the WPS client and connect to the server
from birdy import WPSClient
import birdy
from os import environ
# To display Images from an url
from IPython.core.display import HTML
from IPython.display import Image
# wait until WPS process is finished
import time
[2]:
# This cell is for server admnistration test purpose
# Ignore this cell and modify the following cell according to your needs
fp_server = environ.get('WPS_URL')
print(fp_server) # link to the flyingpigoen server
http://localhost:8093
[3]:
# URL to a flyingpigeon server
# fp_server = 'https://pavics.ouranos.ca/twitcher/ows/proxy/flyginpigeon/wps'
Connect to the server with birdy client
[4]:
# Simple connection (not recommended for larger processing)
fp = WPSClient(fp_server)
# Asyncronous connection with progess status requests
fp_i = WPSClient(url=fp_server, progress=True)
Explore the available processes:
[5]:
# Enter `fp?` for general exploration on processes provided by flyingpigeon
# fp?
# Or get help for a process in particular
help(fp.subset_continents) # or type: fp.subset_countries?
Help on method subset_continents in module birdy.client.base:
subset_continents(resource=None, region='Africa', mosaic=False) method of birdy.client.base.WPSClient instance
Return the data whose grid cells intersect the selected continents for each input dataset.
Parameters
----------
region : {'Africa', 'Asia', 'Australia', 'North America', 'Oceania', 'South America', 'Antarctica', 'Europe'}string
Continent name.
mosaic : boolean
If True, selected regions will be merged into a single geometry.
resource : ComplexData:mimetype:`application/x-netcdf`, :mimetype:`application/x-tar`, :mimetype:`application/zip`
NetCDF Files or archive (tar/zip) containing netCDF files.
Returns
-------
output : ComplexData:mimetype:`application/x-netcdf`
NetCDF output for first resource file.
metalink : ComplexData:mimetype:`application/metalink+xml; version=4.0`
Metalink file with links to all NetCDF outputs.
[11]:
# some test data
# 'https://github.com/roocs/mini-esgf-data/raw/master/test_data/group_workspaces/jasmin2/cp4cds1/data/c3s-cordex/output/EUR-11/IPSL/MOHC-HadGEM2-ES/rcp85/r1i1p1/IPSL-WRF381P/v1/day/psl/v20190212/'
# 'psl_EUR-11_MOHC-HadGEM2-ES_rcp85_r1i1p1_IPSL-WRF381P_v1_day_20060101-20101231.nc'
# Specify input files
base_url = 'https://www.esrl.noaa.gov/psd/thredds/fileServer/Datasets/ncep.reanalysis.dailyavgs/surface/'
urls = [base_url + f'slp.{year}.nc' for year in range(2000, 2002)]
Subset over a bounding box¶
Note that all netCDF input files will be subsetted individually.
[7]:
# bbox subset of one file without interaction to the server
resp = fp.subset_bbox(
resource=urls[0], # can be also a list of files but it is recommended to run large processes with fp_i
lon0=20,
lon1=70,
lat0=10,
lat1=50,
start=None, # optional to select a time periode
end=None,
variable=None, # can be set if the variable is known, otherwise the process detects the variable name
)
# Check the output files:
bbox = resp.get()
[8]:
# bbox of multiple files
resp = fp_i.subset_bbox(
resource=urls,
lon0=20,
lon1=70,
lat0=10,
lat1=50,
start=None, # optional to select a time periode
end=None,
variable=None, # can be set if the variable is known, otherwise the process detects the variable name
)
# using fp_i you need to wait until the processing is complete!
timeout = time.time() + 60*5 # 5 minutes from now
while resp.getStatus() != 'ProcessSucceeded':
time.sleep(1)
if time.time() > timeout: # to avoid endless waiting if the process failed
break
# Get the output files:
bbox = resp.get()
[9]:
# Plot the test file with the flyingpigeon plot function
out_plot = fp.plot_map_timemean(resource=bbox.output)
# and display the output
Image(url= out_plot.get()[0], width=400)
[9]:

[12]:
# If you want to download the result files:
# Download files stored in a metalink
# requires https://github.com/metalink-dev/pymetalink
#
from metalink import download
import tempfile
path = tempfile.mkdtemp()
files = download.get(bbox.metalink, path=path, segmented=False, force=True)
len(files)
Metalink content-type detected.
Downloading to /tmp/tmp_v07l0_1/slp.2000_bbox_subset.nc.
Downloading to /tmp/tmp_v07l0_1/slp.2001_bbox_subset.nc.
[12]:
2
Subset over a continent¶
As for bounding box subsetting, all netCDF input files are subsetted individually. Here, it is possible to specifiy one or multiple continents. If mosaic is True, all continents are merged into one shape before subsetting. If False, each input file is subsetted over each polygon, such that the number of output files is the number of input files times the number of continents.
The subset_countries process works the same with polygons for countries instead of continents.
There are two outputs: * a netCDF file to have a quick test to check if the process went according to the users needs * a metalink file with the list of all output files
[13]:
# Run the process
resp = fp_i.subset_continents(resource=urls, region=['Europe', 'Africa'], mosaic=True)
# using fp_i you need to wait until the processing is complete!
timeout = time.time() + 60*5 # 5 minutes from now
while resp.getStatus() != 'ProcessSucceeded':
time.sleep(1)
if time.time() > timeout: # to avoid endless waiting if the process failed
break
# Check the output files:
cont = resp.get()
[14]:
# plot the test file (url of output) with the flyingpigon plot function
resp = fp.plot_map_timemean(resource=cont.output)
[15]:
# The plot process returnes one graphic file.
# It can be plotted directly by asking birdy to get python objects, instead of links to files.
resp.get(asobj=True).plotout_map
[15]:
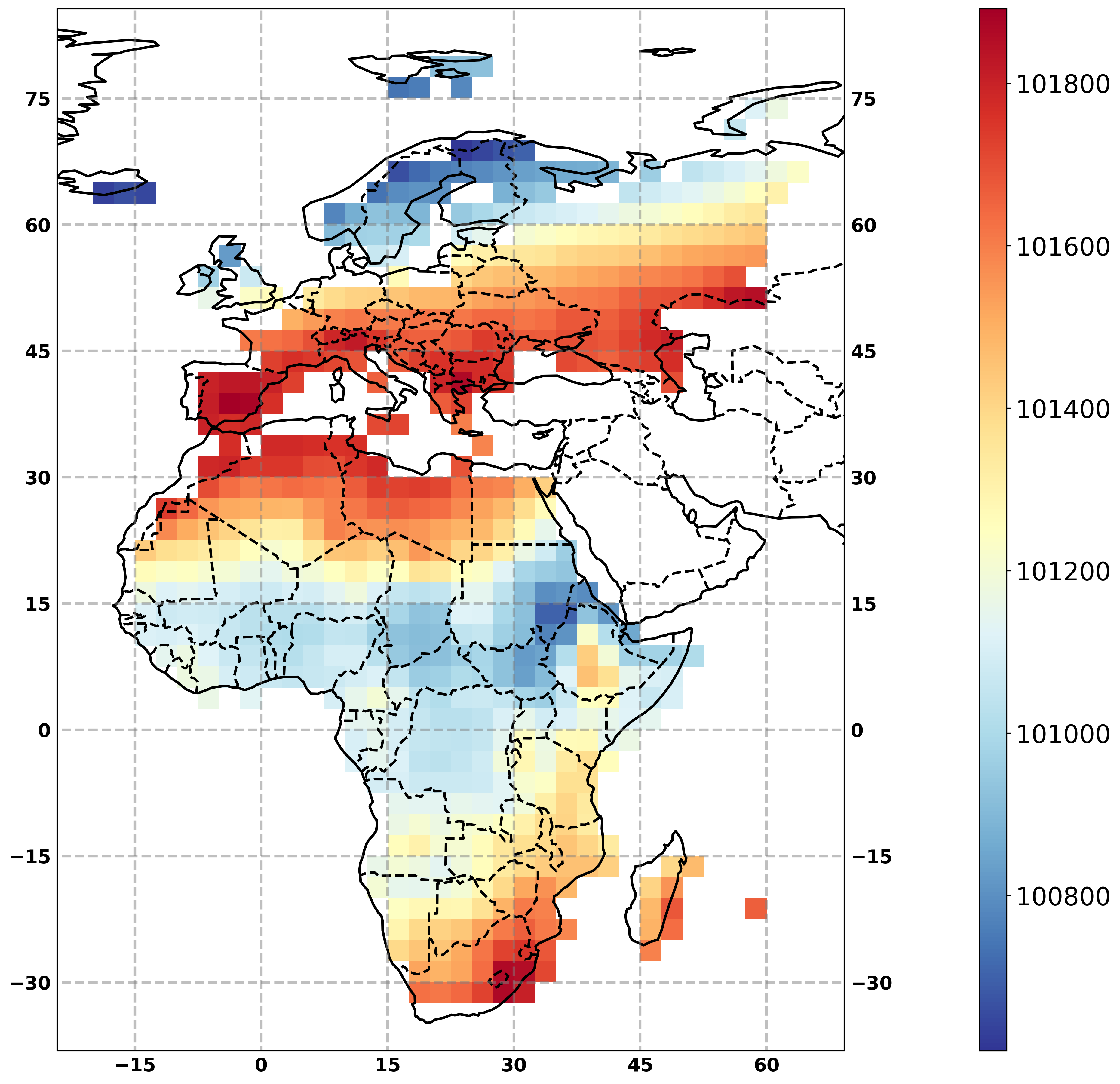
Subset with WFS server¶
[16]:
inputs = dict(nc="https://pavics.ouranos.ca/twitcher/ows/proxy/thredds/dodsC/birdhouse/nrcan"
"/nrcan_northamerica_monthly"
"/tasmax/nrcan_northamerica_monthly_2015_tasmax.nc",
typename="public:USGS_HydroBASINS_lake_na_lev12",
geoserver="https://pavics.ouranos.ca/geoserver/wfs",
fid1="USGS_HydroBASINS_lake_na_lev12.67061",
fid2="USGS_HydroBASINS_lake_na_lev12.67088",
mosaic=False)
# resp = fp_i.subset_wfs_polygon(resource=inputs['nc'],
# typename=inputs['typename'],
# featureids=inputs['fid1'],
# geoserver=inputs['geoserver'],
# mosaic=False,
# start=None,
# end=None,
# variable=None,
# )
[17]:
# # using fp_i you need to wait until the processing is complete!
# timeout = time.time() + 60*5 # 5 minutes from now
# while resp.getStatus() != 'ProcessSucceeded':
# time.sleep(1)
# if time.time() > timeout: # to avoid endless waiting if the process failed
# break
# outputs = resp.get()
[ ]: